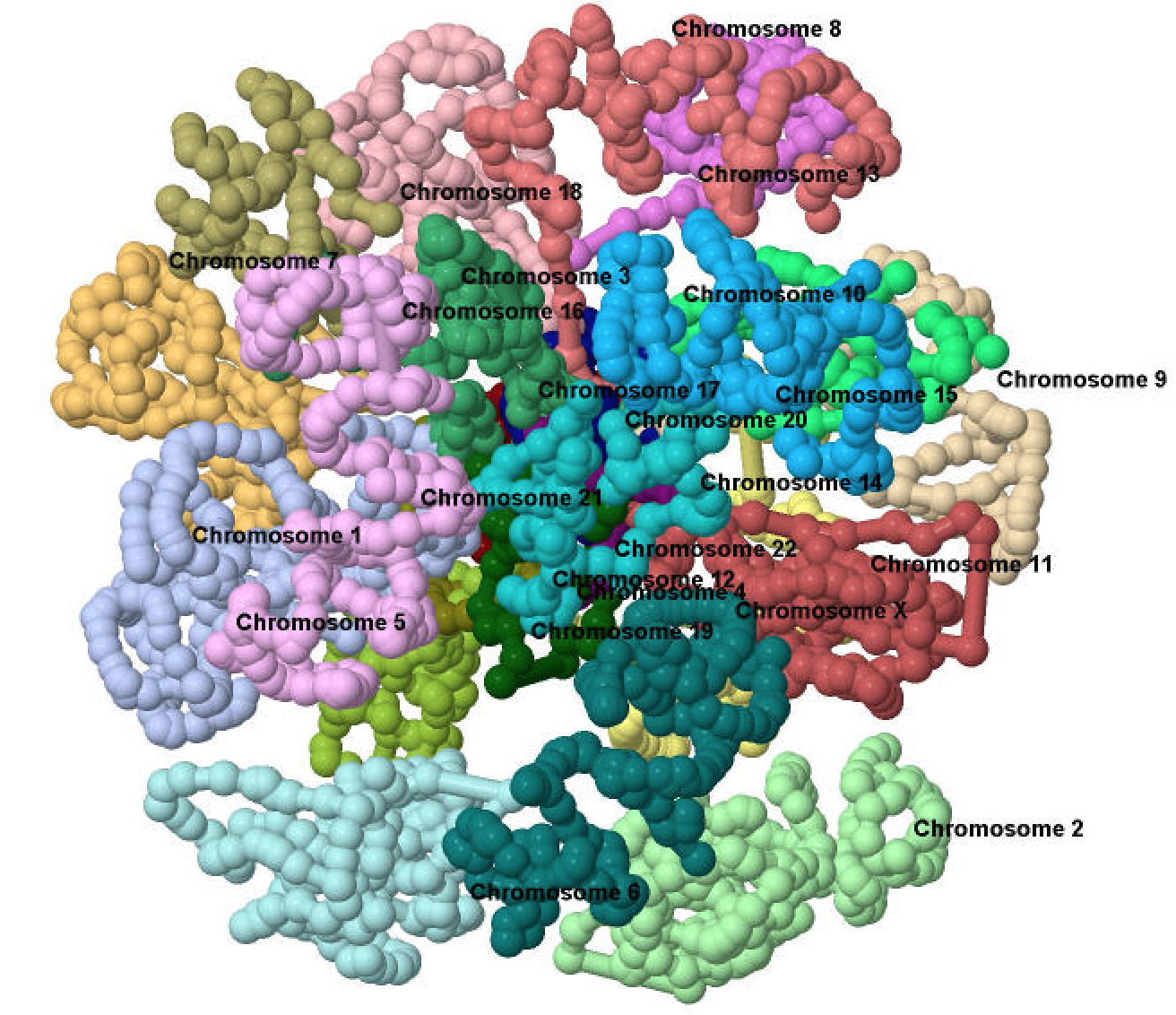
NSF CAREER Project: Analysis, Construction, Visualization, and Modeling of 3D Genome Structures
Journal Publications
1. M. Zhu, X. Deng, T. Joshi, D. Xu, G. Stacey, J. Cheng. Reconstructing Differentially Co-expressed Gene Modules and Regulatory Networks of Soybean Cells. BMC Genomics, 13:434, 2012. [at BMC Genomics web site].
2. Z. Wang, R. Cao, K. Taylor, A. Briley, C. Caldwell, J. Cheng. The Properties of Genome Conformation and Spatial Gene Interaction and Regulation Networks of Normal and Malignant Human Cell Types. PLoS ONE. 8(3):e58793, 2013 [at PLoS ONE's web site].
3. M. Zhu, J. Dahmen, G. Stacey, J. Cheng. Predicting Gene Regulatory Networks of Soybean Nodulation from RNA-Seq Transcriptome Data. BMC Bioinformatics. 14:278, 2013. [at BMC Bioinformatics' website].
4. K.H. Taylor, A. Briley, Z. Wang, J. Cheng, H. Shi, C.W. Caldwell. Aberrant Epigenetic Gene Regulation in Lymphoid Malignancies. Seminars in Hematology. 50(1):38-47, 2013. [at Elsevier's web site].
5. T. Trieu, J. Cheng. Large-scale reconstruction of 3D structures of human chromosomes from chromosomal contact data. Nucleic Acids Research, accepted. [at NAR's website].
6. Q. Qi, J. Li, J. Cheng. Reconstruction of Metabolic Pathways by Combining Probabilistic Graphical Model-based and Knowledge-based Methods. BMC Proceeding, 8(S6):S5, 2014.
7. J. Hou, D. Zhu, J. Cheng (2015). An overview of bioinformatics methods for modeling biological pathways in yeast. Briefings in Functional Genomics. [at BFG]
8. J. Hou, G. Stacey, J. Cheng (2015). Exploring soybean metabolic pathways based on probabilistic graphical model and knowledge-based methods. EURASIP Journal on Bioinformatics and Systems Biology. [at PubMed]
9. J. Li, J. Hou, L. Sun, J.M. Wilkins, Y. Lu, C.E. Niederhuth, B.R. Merideth, T.P. Mawhinney, V. Valeri, C.M. Greenlief, J.C. Walker, W.R. Folk, M. Hannink, D.B. Lubahn, J.A. Birchler, J. Cheng (2015). From Gigabyte to Kilobyte: a Bioinformatics Protocol for Mining Large RNA-Seq Transcriptomics Data. PLoS ONE. 10 (4), e0125000. [at PLoS One]
10. J. Nowotny, S. Ahmed, L. Xu, O. Oluwadare, H. Chen, N. Hensley, T. Trieu, R. Cao, J. Cheng (2015). Iterative reconstruction of three-dimensional models of human chromosomes from chromosomal contact data. BMC Bioinformatics. 16 (1), 338. [at BMC Bioinformatics]
11. R. Cao, J. Cheng (2015). Deciphering the association between gene function and spatial gene-gene interactions in 3D human genome conformation. BMC Genomics. 16 880. [at BMC Genomics]
12. R. Cao, J. Cheng (2016). Integrated Protein Function Prediction by Mining Function Associations, Sequences, and Protein-Protein and Gene-Gene Interaction Networks. Methods. 93:84-91. [at Methods]
13. Y. Jiang et al. An expanded evaluation of protein function prediction methods shows an improvement in accuracy. Genome Biology. 2016.
14. S. Cui, T. Ji, J. Li, J. Cheng, J. Qiu (2016). What if we ignore the random effects when analyzing RNA-seq data in a multifactor experiment?. Statistical Applications in Genetics and Molecular Biology (SAGMB). [at SAGMB]
15. T. Trieu, J. Cheng (2015). MOGEN: a tool for reconstructing 3D models of genomes from chromosomal conformation capturing data. Bioinformatics. [at Bioinformatics]
16. T. Trieu, J. Cheng. 3D Genome Structure Modeling by Lorentzian Objective Function. Nucleic Acids Research, accepted, 2016. [at NAR]
17. B. Adhikari, T. Tuan, J. Cheng. Chromosome3D: Reconstructing Three-Dimensional Chromosomal Structures from Hi-C Interaction Frequency Data using Distance Geometry Simulated Annealing. BMC Genomics, 17:886, 2016. [at BMC Genomics]
18. J. Nowotny, A. Wells, O. Oluwadare, L. Xu, R. Cao, T. Trieu, C. He, and J. Cheng (2016). GMOL: an interactive tool for 3D genome structure visualization. Scientific Reports. 6 (20802). [at Scientific Reports]
19. O. Oluwadare, J. Cheng. ClusterTAD: an unsupervised machine learning approach to detecting topologically associated domains of chromosomes from Hi-C data. BMC Bioinformatics. 18:480, 2017. [at BMC Bioinformatics]
20. X. Rui, J. Wen, A. Quitadamo, J. Cheng, and X. Shi. A deep auto-encoder model for gene expression
prediction. BMC Genomics. 18(S9):845, 2017. [BMC Genomics]
21. J. Lingyan, Y. Wan, J. C. Anderson, J. Hou, S.M. Soliman, J. Cheng, S.C. Peck. Genetic Dissection of
Arabidopsis MAP Kinase Phosphatase 1 (AtMKP1)-dependent PAMP-induced transcription pathways. Journal
of Experimental Botany. 68(18):5207-5220, 2017. [at JEB]
22. O. Oluwadare; Y. Zhang; J. Cheng. A maximum likelihood algorithm for reconstructing 3D structures of human
chromosomes from chromosomal contact data, BMC Genomics , 19:161, 2018. [at BMC Genomics]
23. O. Oluwadare, Max Highsmith, J. Cheng. An overview of methods for reconstructing 3D chromosome and genome structures from Hi-C data. Biological Procedures Online. Accepted.
24. T. Trieu, O. Oluwadare, J. Cheng. Hierarchical Reconstruction of High-Resolution 3D Models of Human Chromosomes. Scientific Reports, 2019.
25. T. Trieu, O. Oluwadare, J. Wopata, and J. Cheng. GenomeFlow: A Comprehensive Graphical Tool for Modeling and Analyzing 3D Genome Structure. Bioinformatics, accepted.
Conference Abstracts, Posters, and Presentations
1. Z. Wang, R. Cao, K. Taylor, A. Briley, C. Caldwell, J. Cheng. The Properties of Human Genome Conformation and Spatial Gene Interaction and Regulation Networks . Pacific Symposium on Biocomputing , Hawaii, 2013. (Poster presentation)
2. T. Trieu, J. Cheng. Consructing Three-Dimensional Structures of Human Chromosomes from Chromosmal Contact Data. Mid-South Computational Biology and Bioinformatics Conference, Columbia, MO, 2013. (Oral presentation).
3. S. Ahmed, J. Cheng. Iterative Reconstruction of Three-Dimensinal Structures of Human Genome from Chromosomal Contact Data. Mid-South Computational Biology and Bioinformatics Conference, Columbia, MO, 2013. (Poster presentation)
4. C. He, A.O. Wells, J. Cheng. GMol: A Tool for 3D Genome Structure Visualization. Mid-South Computational Biology and Bioinformatics Conference, Columbia, MO, 2013. (Poster presentation)
5. S. Ahmed, J. Cheng. Iterative Reconstruction of Three-Dimensinal Structures of Human Genome from Chromosomal Contact Data. Life Science Week Symposium, University of Missouri, Columbia, MO, 2013. (Poster presentation)
6. Z. Wang, R. Cao, K. Taylor, A. Briley, C. Caldwell, J. Cheng. The Properties of Human Genome Conformation and Spatial Gene Interaction and Regulation Networks . Mid-South Computational Biology and Bioinformatics Conference, Columbia, MO, 2013. (Oral presentation, won the first place award in the post-doctoral presentation category).
7. C. He, A.O. Wells, J. Cheng. GMol: A Tool for 3D Genome Structure Visualization. The Great Lake Bioinformatics Conference, Puttsburg, PA, 2013.(Poster presentation)
8. T. Trieu, J. Cheng. Consructing Three-Dimensional Structures of Human Chromosomes from Chromosmal Contact Data. Life Science Week, University of Missouri, Columbia, MO, 2013. (Poster presentation).
9. R. Cao, J. Cheng. Deciphering the Association between Gene Function and Spatial Gene-Gene Interactions in 3D Genome Conformation. The 21st Annual International Conference on Intelligent Systems for Molecular Biology (ISMB). Berlin, Germany, 2013.
10. R. Cao, J. Cheng. Deciphering the Association between Gene Function and Spatial Gene-Gene Interactions in 3D Genome Conformation. The Automated Protein Function Annotation (AFA) SIG meeting of the 21st Annual International Conference on Intelligent Systems for Molecular Biology. Berlin, Germany, 2013.
11. R. Cao, J. Cheng (2015). Integrated Protein Function Prediction by Mining Protein Sequences, Function Associations, Protein Interaction Networks, and Gene Interaction Networks. AFA SIG meeting, Intelligent Systems for Molecular Biology (ISMB). Dublin, Ireland.
12. J. Nowotny, S. Ahmed, L. Xu, N. Hensley, J. Cheng (2015). Iterative Reconstruction of Three-Dimensional Models of Human Chromosomes from Chromosomal Contact Data (poster presentation). Undergraduate Research and Creativity Forum. University of Missouri, Columbia.
13. J. Nowotny, S. Ahmed, L. Xu, N. Hensley, J. Che (2015). Iterative Reconstruction of Three-Dimensional Models of Human Chromosomes from Chromosomal Contact Data (poster presentation). MU Life Science Week. University of Missouri, Columbia.
14. O. Oluwadare, J. Nowotny, S. Ahmed, L. Xu, H. Chen, N. Hensley, T. Trieu, R. Cao, J. Cheng (2016). Iterative reconstruction of three-dimensional models of human chromosomes from chromosomal contact data. The Mid-South Computational Biology and Bioinformatics Society Conference (MCBIOS). Memphis, TN.
15. T. Trieu, J. Cheng (2015). Reconstructing 3D Model of the Entire Human Genome from Inter- and Intra-Chromosomal Contacts (poster presentation). MU Life Science Week. University of Missouri, Columbia.
16. R. Cao, J. Cheng (2015). The Association Between Gene Function and Spatial Gene-Gene Interactions in 3D Human Genome Conformation (poster presentation). MU Life Science Week. University of Missouri, Columbia.
17. J. Nowotny, J. Cheng (2016). A computational approach to construct three-dimensional models of human chromosomes. The Undergraduate Research Forum. University of Missouri, Columbia.
18. J. Cheng (2017). 3D Genome Structure Modeling by Lorentzian Objective Function (highlight presentation). The 8th ACM Conference on Bioinformatics, Computational Biology, and Health Informatics (ACM BCB), Boston, MA.[PPT slides]
Other Presentations of Genome Structure Modeling
1. J. Cheng. Analysis and Reconstruction of 3D Conformation of Human Genome. The Great Lake Bioinformatics Conference, Cincinati, OH, 2014.
2. J. Cheng. The Properties of 3D Human Genome. The ACM BCB Conference, Washington DC, Sept., 2013.
3. J. Cheng. Computational Modeling of 2D and 3D Human Genome Structure. Purdue University, April, 2013.
4. J. Cheng. Computational Construction, Analysis and Application of Biological Networks. Wayne State University, Nov., 2012.
5. J. Cheng. Modeling of 3D Human Genome Conformation, the Jackson Laboratory, Connecticuit, 2015.
6. J. Cheng. Modeling of 3D Human Genome Structure and its Application, Department of Biostatistics, Virginia Commonwealth University, 2016.
7. J. Cheng. Modeling 3D Human Genome Structures, School of Medicine, Indiana University, 2017. [PPT slides]
8. J. Cheng. Data-driven modeling of protein structure, 3D genome, and gene regulatory networks, Danforth Plant Science Center, 2018.
9. J. Cheng. 3D Genome Modeling, Michigan State University, 2019.
Theses & Dissertations
1. Zheng Wang. Revealing the Conformation and Properties of Human Genome, Protein Molecules, and Protein Domain Co-Occurrence Network. PhD Dissertation. University of Missouri, Columbia, 2012.
2. Sharif Ahmed. Iterative reconstruction of three-dimensional model of human genome from chromosomal contact data (final version). (2014). University of Missouri - Columbia.
3. Y. Zhang. EM Algorithm for Reconstructing 3D Structures of Human Chromosomes from Chromosomal Contact Data. Master's Thesis. University of Missouri - Columbia, 2016.
4. R. Cao. Genome Data Analysis, Protein Structure and Function Predictioin by Machine Learning Techniques. PhD Dissertation. University of Missouri - Columbia, 2016.
5. Tuan Trieu. T. Trieu. 3D Genome Structure Reconstruction from Chromosomal Contact Data. PhD Dissertation. University
of Missouri - Columbia, 2017
6. O. Oluwadare. Data Mining and Machine Learning Methods for Chromosome Conformation Data Analysis. PhD Dissertation. University of Missouri – Columbia, 2019.
The Hi-C data of three cells / cell lines (MHH-CALL4-CELL-LINE, Primary ALL B Cell, RL Cell Line published in Wang et al., PLoS ONE, 2013.
The 3D models of 23 pairs of chromosomes of healthy and luekemia B-cells constructed from Hi-C data published in Trieu and Cheng, Nucleic Acids Research, 2014.
Open source GMOL tool for visualizing 3D genome structures
Gene3D: an open source tool for reconstructing 3D genome structure from chromosomal conformation capturing (e.g. Hi-C) data.
3D genome models of healthy and cancerous B-cells
MOGEN tool for genome structure modeling
Chromosome3D tool for chromosome structure modeling
LorDG: Lorentzian3DGenome tool for genome structure modeling
ClusterTAD for identifying topologically associated domains (TAD) from Hi-C data
3DMax: a maximum likelihood approach to chromosome structure modeling
GenomeFlow
Hierarchical3DGenome
DeepChroMap: deep learning to remove noise in chromosomal contact data
Genome Structure Database (GSDB)
GSDB at GitHub
Courses
Computational Modeling of Molecular Structures (Spring, 2013; Spring, 2014; Spring, 2016, Spring 2018)
Computational Optimization Methods (Fall, 2013; Fall 2014)
Videos and Gallery
The movie of folding / modeling Chromosome 11 of a human B-cell posted at YouTube.
The video of modeling a 3D human chromosome structure using a contact-driven gradient descent method. (Publically demonstrated first on March 14 to K1-12 students during the 2013 College of Enginering E-Week). The video was composed by Trieu Tuan, a PhD student in the Computer Science Department.
1. The E-Week Lab Exhibit for Columbia K1-12 Students, Computational Modeling of Genome Strucutres, College of Engineering, University of Missouri, Columbia, March, 2013.
2. Training a high school student intern (Charles Shang) in the summer, 2012. His work in our group won semi-finalist in 2012 Siemens Science Talent Search and 2013 Intel Science Talent Search.
3. The video recording Avery Well's presentation to K12 students during 2014 College of Engineering's E-Week Exhibit at the University of Missouri, Columbia.
4. Presentation to K12 students during 2015 College of Engineering's E-Week at the University of Missouri, Columbia.
5. Hannah Chen, a high school intern in our lab won 2015 semi-finalist of Siemens Science Talent Search.
6. Richard Liu, a high school student won regional scientific competition, 2016.
7. Julia Wopata, an undergraduate student, joined us to work on 3D genome modeling, 2017-2018.
8. Jena', African American REU undergraduate student.
News
Our high resolution 3D genome modeling work is reported by MU News Burea
Prof. Jianlin Cheng was honored as one of MU Chancellor's top achievers because of his work on this CAREEER project.
The research was featured in the Mizzou Alumni Magazine, 2013.
Cheng takes aim at genetic mysteries
Cheng receives NSF CAREER Award
Charles Shang Won Semi-Finalist of 2012 Simmens Science Talent Search
Charles Shang Won Semi-Finalist of 2013 Intel Science Talent Search
People
Principle Investigator
Dr. Jianlin Cheng
Graduate Students
Sharif Ahmed (graduated), Renzhi Cao (graduated), Chenfeng He (transferred), Tuan Anh Trieu (graduated), Dr. Zheng Wang (graduated in 2012), Lingfei Xu (graduated), Adhari Badri (graduated), Rajani Kanmanthareddy (graduated), Oluwatosin Oluwadare, Yuxiang Zhang (graduated), Avery Wells, Tianshu Wei (graduated)
Undergraduate Students
Avery Owen Wells, Jackson Nowotny, Sarah Kang, Hannah Wiedner, Nicholas Hunkins, Noelan Hensley, Julia Wopata, Jena'
High School Students
Charles Shang, Angela Zhang, Richard Liu, Hannah Chen
Contact
Director of Bioinformatics, Data Mining and Machine Learning Laboratory
Professor
Department of Computer Science
Informatics Institute
C. Bond Life Science Center
College of Engineering
University of Missouri, Columbia, MO 65211-2060
Primary Office: EBW 109
Lab: EBN 226, 304, 305, 307
Phone: 573-882-7306
Fax: 573-882-8318
Email: chengji@missouri.edu

